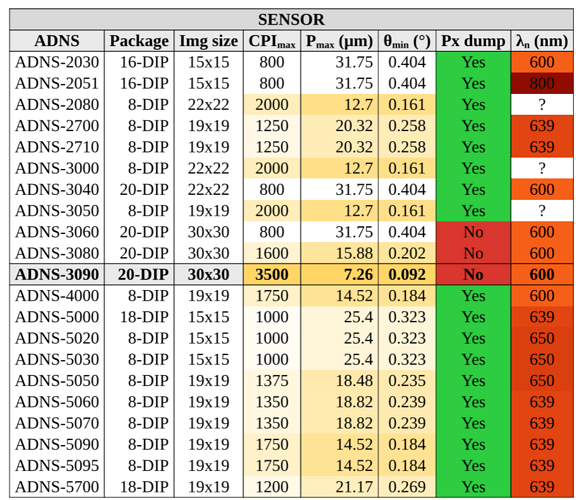
After a lot of work I made a heatmap table, but I’m sure it can be improved.
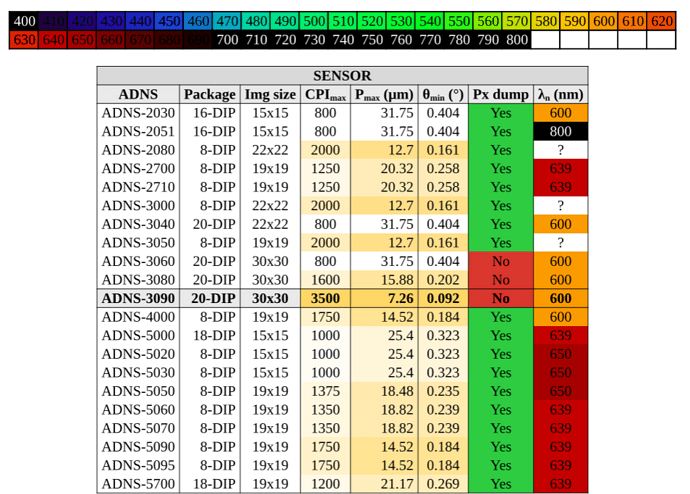
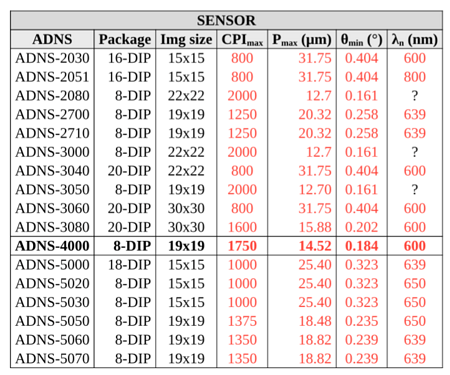
My goal was to replicate the “conditional formatting” from google sheets. Here is the table I wish to reproduce (note that the column PX dump has manual coloring):
First thing I did was to recreate the yellow gradient:
#let gsheet = gradient.linear(
rgb("ffffff"),
rgb("fff5d9"),
rgb("ffebb3"),
rgb("ffe18d"),
rgb("ffd666"),
)
After a lot of searching, I found that you could sample a gradient at 0-100%, so I made a normalization function to normalize the cell value between 0 and 100%. In the case of the CPI_max column, min would be 800 and max would be 3500.
#let normalize(value, min, max) = {
let n = (value - min) / (max - min) * 100
n = calc.clamp(n, 0, 100)
return n * 1%
}
And here comes the clunky part. I tried with dict but I couldn’t make it work. Since I wish to support the std color maps, I included them in the function:
#let colmap(map, value, min, max) = {
let c = normalize(value, min, max)
if map == "turbo" {return gradient.linear(..color.map.turbo).sample(c)}
else if map == "cividis" {return gradient.linear(..color.map.cividis).sample(c)}
else if map == "rainbow" {return gradient.linear(..color.map.rainbow).sample(c)}
else if map == "spectral" {return gradient.linear(..color.map.spectral).sample(c)}
else if map == "viridis" {return gradient.linear(..color.map.viridis).sample(c)}
else if map == "inferno" {return gradient.linear(..color.map.inferno).sample(c)}
else if map == "magma" {return gradient.linear(..color.map.magma).sample(c)}
else if map == "plasma" {return gradient.linear(..color.map.plasma).sample(c)}
else if map == "rocket" {return gradient.linear(..color.map.rocket).sample(c)}
else if map == "mako" {return gradient.linear(..color.map.mako).sample(c)}
else if map == "vlag" {return gradient.linear(..color.map.vlag).sample(c)}
else if map == "icefire" {return gradient.linear(..color.map.icefire).sample(c)}
else if map == "flare" {return gradient.linear(..color.map.flare).sample(c)}
else if map == "crest" {return gradient.linear(..color.map.crest).sample(c)}
else if map == "gsheet" {return gsheet.sample(c)}
}
And finally the table:
#import table: cell, header, hline
#let yes = cell(fill: green)[Yes]
#let no = cell(fill: red.darken(15%))[No]
#let cpi(val) = cell(fill: colmap("gsheet", val, 800, 3500))[#val]
#let pm(val) = cell(fill: colmap("gsheet", val, 31.75, 7.26))[#val]
#let tm(val) = cell(fill: colmap("gsheet", val, 0.404, 0.092))[#val]
#let Ln(val) = cell(fill: colmap("turbo", val, -200, 800))[#val]
#set table(
fill: (x, y) =>
if y == 0 { gray.lighten(55%) }
else if y == 1 { gray.lighten(75%) },
align: (x, y) => (
if ((x == 1) or (x == 4)) and (y > 1) { right }
else { center }
)
)
#show table.cell.where(y: 0): set text(weight: "bold")
#show table.cell.where(y: 1): set text(weight: "bold")
#show table.cell.where(y: 12): set text(weight: "bold")
#table(
columns: 8,
align: center + horizon,
stroke: (x: linestrokewidth, y: none),
hline(y: 0, stroke: (dash: "solid", thickness: linestrokewidth),),
hline(y: 1, stroke: (dash: "solid", thickness: linestrokewidth),),
hline(y: 2, stroke: (dash: "solid", thickness: linestrokewidth),),
hline(y: 12, stroke: (dash: "solid", thickness: linestrokewidth),),
hline(y: 13, stroke: (dash: "solid", thickness: linestrokewidth),),
hline(y: 23, stroke: (dash: "solid", thickness: linestrokewidth),),
// Header
header(
cell(colspan: 8)[SENSOR],
[ADNS], [Package], [Img size], [CPI#sub[max]], [P#sub[max] (#sym.mu\m)], [#sym.theta#sub[min] (#sym.degree)], [Px dump], [#sym.lambda#sub[n] (nm)],
),
// Data
[ADNS-2030], [16-DIP], [15x15], cpi(800), pm(31.75), tm(0.404), yes, Ln(600),
[ADNS-2051], [16-DIP], [15x15], cpi(800), pm(31.75), tm(0.404), yes, Ln(800),
[ADNS-2080], [8-DIP], [22x22], cpi(2000), pm( 12.7), tm(0.161), yes, [?],
[ADNS-2700], [8-DIP], [19x19], cpi(1250), pm(20.32), tm(0.258), yes, Ln(639),
[ADNS-2710], [8-DIP], [19x19], cpi(1250), pm(20.32), tm(0.258), yes, Ln(639),
[ADNS-3000], [8-DIP], [22x22], cpi(2000), pm( 12.7), tm(0.161), yes, [?],
[ADNS-3040], [20-DIP], [22x22], cpi(800), pm(31.75), tm(0.404), yes, Ln(600),
[ADNS-3050], [8-DIP], [19x19], cpi(2000), pm( 12.7), tm(0.161), yes, [?],
[ADNS-3060], [20-DIP], [30x30], cpi(800), pm(31.75), tm(0.404), no, Ln(600),
[ADNS-3080], [20-DIP], [30x30], cpi(1600), pm(15.88), tm(0.202), no, Ln(600),
cell(fill: gray.lighten(75%))[ADNS-3090], cell(fill: gray.lighten(75%))[20-DIP], cell(fill: gray.lighten(75%))[30x30], cpi(3500), pm(7.26), tm(0.092), no, Ln(600),
[ADNS-4000], [8-DIP], [19x19], cpi(1750), pm(14.52), tm(0.184), yes, Ln(600),
[ADNS-5000], [18-DIP], [15x15], cpi(1000), pm( 25.4), tm(0.323), yes, Ln(639),
[ADNS-5020], [8-DIP], [15x15], cpi(1000), pm( 25.4), tm(0.323), yes, Ln(650),
[ADNS-5030], [8-DIP], [15x15], cpi(1000), pm( 25.4), tm(0.323), yes, Ln(650),
[ADNS-5050], [8-DIP], [19x19], cpi(1375), pm(18.48), tm(0.235), yes, Ln(650),
[ADNS-5060], [8-DIP], [19x19], cpi(1350), pm(18.82), tm(0.239), yes, Ln(639),
[ADNS-5070], [8-DIP], [19x19], cpi(1350), pm(18.82), tm(0.239), yes, Ln(639),
[ADNS-5090], [8-DIP], [19x19], cpi(1750), pm(14.52), tm(0.184), yes, Ln(639),
[ADNS-5095], [8-DIP], [19x19], cpi(1750), pm(14.52), tm(0.184), yes, Ln(639),
[ADNS-5700], [18-DIP], [19x19], cpi(1200), pm(21.17), tm(0.269), yes, Ln(639),
))
I think the way I did it is very bad, but I couldn’t find any resource online… The best would be a show rule or a set rule that applies directly on a column, something like this:
#set table(
fill: (x, y) =>
if x == 4 {colmap("gsheet", table.cell.body, 800, 3500)}
)
But I don’t know how to do that.
Also, the λ column is supposed to convey the color at that specific wavelength, but the normalization 400-800nm (infrared to ultraviolet) doesn’t work well on the std color maps like rainbow or turbo